This page was generated from unit-5.5-cuda/poisson_cuda.ipynb.
5.5.1 Solving the Poisson Equation on devices¶
After finite element discretization we obtain a (linear) system of equations.
The new ngscuda module moves the linear operators to the Cuda - decice.
The host is stearing, data stays on the device
The module is now included in NGSolve Linux - distributions, and can be used whenever an accelerator card by NVIDIA is available, and the cuda-runtime is installed.
[1]:
from ngsolve import *
from ngsolve.webgui import Draw
from time import time
[2]:
mesh = Mesh(unit_square.GenerateMesh(maxh=0.1))
for l in range(5): mesh.Refine()
fes = H1(mesh, order=2, dirichlet=".*")
print ("ndof =", fes.ndof)
u, v = fes.TnT()
with TaskManager():
a = BilinearForm(grad(u)*grad(v)*dx+u*v*dx).Assemble()
f = LinearForm(x*v*dx).Assemble()
gfu = GridFunction(fes)
jac = a.mat.CreateSmoother(fes.FreeDofs())
with TaskManager():
inv_host = CGSolver(a.mat, jac, maxiter=2000)
ts = time()
gfu.vec.data = inv_host * f.vec
te = time()
print ("steps =", inv_host.GetSteps(), ", time =", te-ts)
# Draw (gfu);
ndof = 472321
steps = 1738 , time = 9.114434003829956
Now we import the NGSolve - cuda library.
It provides
an
UnifiedVector, which allocates memory on both, host and device. The data is updated on demand either on host, or on device.NGSolve - matrices can create their counterparts on the device. In the following, the conjugate gradients iteration runs on the host, but all operations involving big data are performed on the accelerator.
[3]:
try:
from ngsolve.ngscuda import *
except:
print ("no CUDA library or device available, using replacement types on host")
ngsglobals.msg_level=1
fdev = f.vec.CreateDeviceVector(copy=True)
no CUDA library or device available, using replacement types on host
No device creator function, creating host vector
[4]:
adev = a.mat.CreateDeviceMatrix()
jacdev = jac.CreateDeviceMatrix()
inv = CGSolver(adev, jacdev, maxsteps=2000, printrates=False)
ts = time()
res = (inv * fdev).Evaluate()
te = time()
print ("Time on device:", te-ts)
diff = Norm(gfu.vec - res)
print ("diff = ", diff)
Time on device: 29.23365807533264
diff = 0.0
On an A-100 device I got (for 5 levels of refinement, ndof=476417):
Time on device: 0.4084775447845459 diff = 3.406979028373306e-12CG Solver with Block-Jacobi and exact low-order solver:¶
Additive Schwarz preconditioner:
with
\(P\) .. embedding of low-order space
\(A_i\) .. blocks on edges/faces/cells
\(E_i\) .. embedding matrices
[5]:
fes = H1(mesh, order=5, dirichlet=".*")
print ("ndof =", fes.ndof)
u, v = fes.TnT()
with TaskManager():
a = BilinearForm(grad(u)*grad(v)*dx+u*v*dx).Assemble()
f = LinearForm(x*v*dx).Assemble()
gfu = GridFunction(fes)
jac = a.mat.CreateBlockSmoother(fes.CreateSmoothingBlocks())
lospace = fes.lospace
loinv = a.loform.mat.Inverse(inverse="sparsecholesky", freedofs=lospace.FreeDofs())
loemb = fes.loembedding
pre = jac + loemb@loinv@loemb.T
print ("mat", a.mat.GetOperatorInfo())
print ("preconditioner:")
print(pre.GetOperatorInfo())
with TaskManager():
inv = CGSolver(a.mat, pre, maxsteps=2000, printrates=False)
ts = time()
gfu.vec.data = inv * f.vec
te = time()
print ("iterations =", inv.GetSteps(), "time =", te-ts)
ndof = 2947201
mat SparseMatrixd (nze=91286401), h = 2947201, w = 2947201
preconditioner:
SumMatrix, h = 2947201, w = 2947201
BlockJacobi-d, h = 2947201, w = 2947201
EmbeddedTransposeMatrix, h = 2947201, w = 2947201
EmbeddedMatrix, h = 2947201, w = 118401
SparseCholesky-d, h = 118401, w = 118401
iterations = 37 time = 5.254413843154907
[6]:
adev = a.mat.CreateDeviceMatrix()
predev = pre.CreateDeviceMatrix()
fdev = f.vec.CreateDeviceVector()
with TaskManager():
inv = CGSolver(adev, predev, maxsteps=2000, printrates=False)
ts = time()
gfu.vec.data = (inv * fdev).Evaluate()
te = time()
print ("iterations =", inv.GetSteps(), "time =", te-ts)
No device creator function, creating host vector
iterations = 37 time = 5.253296375274658
on the A-100:
SumMatrix, h = 2896001, w = 2896001 N4ngla20DevBlockJacobiMatrixE, h = 2896001, w = 2896001 EmbeddedTransposeMatrix, h = 2896001, w = 2896001 EmbeddedMatrix, h = 2896001, w = 116353 N4ngla17DevSparseCholeskyE, h = 116353, w = 116353 iterations = 37 time= 0.6766986846923828Using the BDDC preconditioner:¶
For the BDDC (balancing domain decomposition with constraints) preconditioning, we build a FEM system with relaxed connectivity:
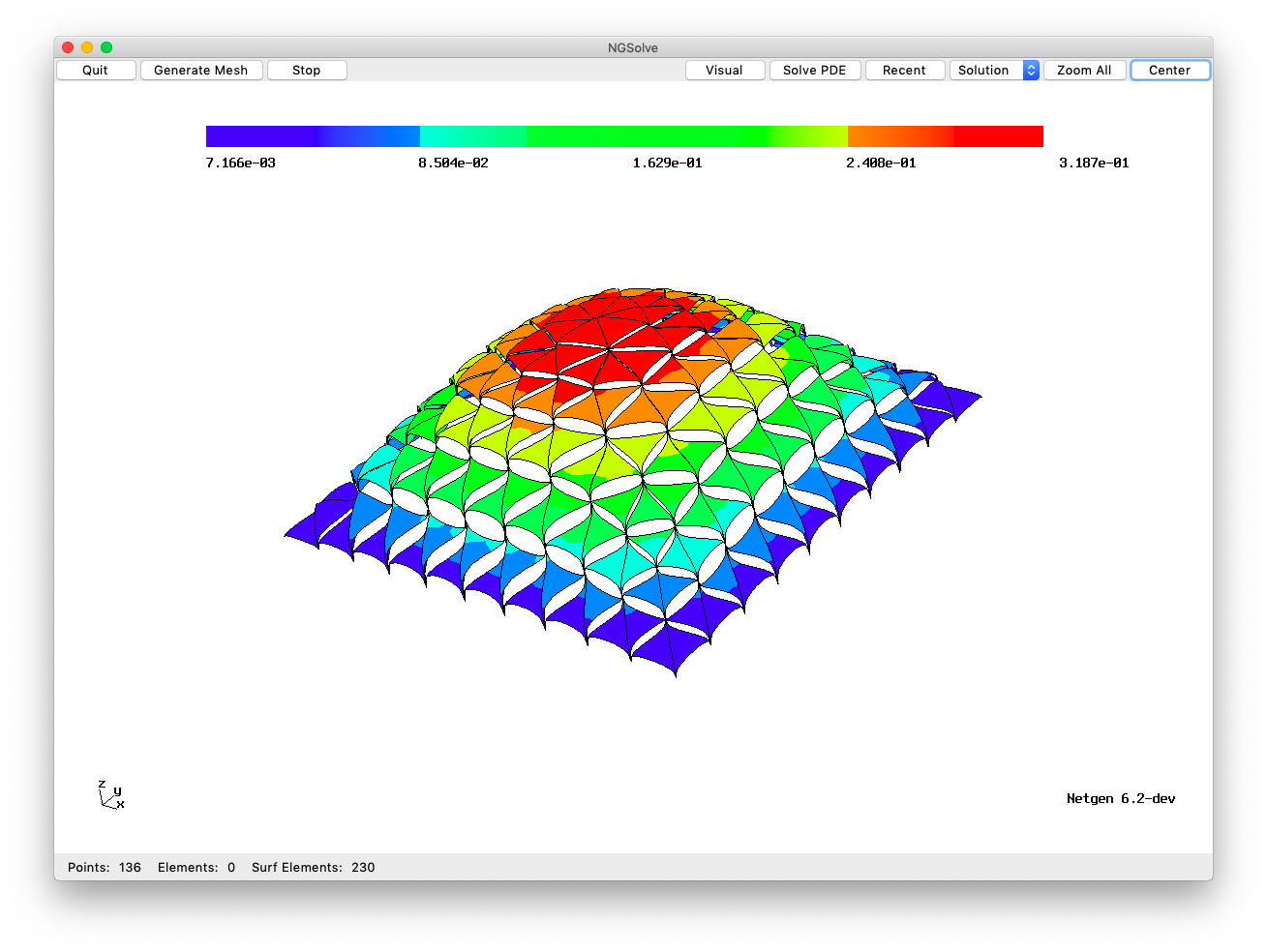
This allows for static condensation of all local and interface degrees of freedom, only the wirebasket dofs enter the global solver. The resulting matrix \(\tilde A\) is much cheaper to invert.
The preconditioner is
with an averagingn operator \(R\).
[7]:
mesh = Mesh(unit_square.GenerateMesh(maxh=0.1))
for l in range(3): mesh.Refine()
fes = H1(mesh, order=10, dirichlet=".*")
print ("ndof =", fes.ndof)
u, v = fes.TnT()
with TaskManager():
a = BilinearForm(grad(u)*grad(v)*dx+u*v*dx)
pre = Preconditioner(a, "bddc")
a.Assemble()
f = LinearForm(x*v*dx).Assemble()
gfu = GridFunction(fes)
with TaskManager():
inv = CGSolver(a.mat, pre, maxsteps=2000, printrates=False)
ts = time()
gfu.vec.data = (inv * f.vec).Evaluate()
te = time()
print ("iterations =", inv.GetSteps(), "time =", te-ts)
Mesh bisection
Bisection done
Mesh bisection
Bisection done
Mesh bisection
Bisection done
ndof = 737601
iterations = 52 time = 3.389190435409546
[8]:
predev = pre.mat.CreateDeviceMatrix()
print (predev.GetOperatorInfo())
SumMatrix, h = 737601, w = 737601
ProductMatrix, h = 737601, w = 737601
ProductMatrix, h = 737601, w = 737601
SparseMatrixd (nze=2328408), h = 737601, w = 737601
SparseCholesky-d, h = 737601, w = 737601
SparseMatrixd (nze=2328408), h = 737601, w = 737601
SparseMatrixd (nze=56311200), h = 737601, w = 737601
[9]:
adev = a.mat.CreateDeviceMatrix()
predev = pre.mat.CreateDeviceMatrix()
fdev = f.vec.CreateDeviceVector()
with TaskManager():
inv = CGSolver(adev, predev, maxsteps=2000, printrates=False)
gfu.vec.data = (inv * fdev).Evaluate()
print ("iterations =", inv.GetSteps())
No device creator function, creating host vector
iterations = 52
Vite - traces:
[ ]: